
At work we recently got Databricks which utilizes open source technologies under the hood. This got me thinking whether I could create a Databricks equivalent with open source software in my homelab. Over the Thanksgiving holiday week, I started playing around with deploying JupyterHub, Minio, and Spark on Kubernetes with Helm. I was able to get a working proof of concept (PoC) that would allow me to read raw log data from Minio using Spark jobs initiated by Python Jupyter notebook to ingest those events into a Spark schema, write that data as a Delta table, and then query said Delta table using a Jupyter notebook.
Goals
- Minio running on k8s
- Jupyterhub running on k8s
- Apache Spark running on k8s
- Ability to submit Spark jobs from Jupyter notebooks remotely
- Ingest data from Minio
- Write data as a Delta table to Minio
- Query Delta table with a Jupyter notebook
Background
What is Apache Spark?
Apache Spark is an open source analytics engine used for big data workloads. It can handle both batches as well as real-time analytics and data processing workloads. Apache Spark started in 2009 as a research project at the University of California, Berkeley.
What is JupyterHub?
JupyterHub is an open source tool that lets you host a distributed Jupyter Notebook environment. With JupyterHub, users can log in to the server, and write Python code in a web browser, without having to install software on their local machine.
What is Minio?
MinIO is a high-performance, S3 compatible object store. It is built for large scale AI/ML, data lake and database workloads. It is software-defined and runs on any cloud or on-premises infrastructure.
What is Helm?
Helm is a tool that automates the creation, packaging, configuration, and deployment of Kubernetes applications by combining your configuration files into a single reusable package. Helm provides one of the most accessible solutions to this problem, making deployments more consistent, repeatable, and reliable. To simplify this concept, think of Helm charts as a blueprint to building a house. The real magic is the ability to tune the blueprint such as make the rooms bigger, add a sunroom, add a home security system, or add furniture to a room. Thus there is no need to know how to do these things but rather choosing what you want.
To provide a real world example, Gitlab has a Helm chart. You have the ability to configure the database as a single instance vs. high availability, the database password, how many instances of Gitlab to run concurrently for high availability, what ingress controller to use, and more. I don’t have any idea how to run Postgres in high availability mode on k8s but the Helm chart knows how to configure that. Thus allowing me to choose what I want but no need to know how.
What is Kubernetes?
Kubernetes (k8s) is an open-source container-orchestration system for automating application deployment, scaling, and management. It was originally designed by Google, and is now maintained by the Cloud Native Computing Foundation. It aims to provide a “platform for automating deployment, scaling, and operations of application containers across clusters of hosts”. It works with a range of container tools, including Docker. Many cloud services offer a Kubernetes-based platform or infrastructure as a service (PaaS or IaaS) on which Kubernetes can be deployed as a platform-providing service. Many vendors also provide their own branded Kubernetes distributions.
What is Delta tables?
Delta Tables is a cutting-edge cloud storage technology that makes storing and managing large volumes of data easy. It provides optimized performance for analytics workloads, making it an ideal choice for any business looking to maximize the value of its data. With Delta Tables, you can easily ingest and process your data in real time, allowing for faster access to insights and analytics. Additionally, Delta Tables offers advanced features such as ACID transactions, time travel capabilities, and integrated file management.
Network digram

Dependencies: The nine layers of Hell
Let’s talk about dependencies (deps) for a moment, more specially the Apache Spark version and Spark library versions. If you don’t use the proper versions of dependencies you can receive all sorts of issues, including errors that are red herrings. First, start by choosing a version of Apache Spark to work with, for this blog post we will use 3.5.0. Next, we need to ensure that Jupyterhub has released a new Docker image for that version of Spark. Next, we will need to download the following JAVA libraries: hadoop-aws, aws-java-sdk-core, aws-java-sdk-s3, aws-java-sdk-dynamodb, delta-spark, and delta-storage. These JAVA dependencies (JARs) will be injected into our pyspark context in our Jupyter notebook to expand it’s capabilities. Lastly, we will need to install Python dependencies such as pyspark, minio, and delta-spark.
Example: hadoop-aws
I would like to take the time to walk through a practical example of grabbing the correct dependency. For this example, we are going to grab hadoop-aws. Follow this hyperlink for hadoop-aws, start by selecting a new version, at the time of writing this the newest version is 3.3.6. Scroll down to the “Provided Dependencies” section and review the listed deps.

Next, run the following Docker command: docker run -it jupyter/pyspark-notebook:x86_64-spark-<spark version> ls /usr/local/spark/jars/ | grep 'hadoop-' to determine the version of hadoop-client, which represents your hadoop-common version.

For this blog post, all dependencies related to Spark, it’s important to ensure you choose the dependencies that match your hadoop-common version. Recursively loop through the list of hadoop-aws versions until we find one that matches our version. Lastly, you need to review the “Compile Dependencies” for additional dependencies. Depending on the dependency you may need to recursively go look at its “Provided Dependencies” and “Compile Dependencies”. The hadoop-aws dependency has a dependency on aws-java-sdk-bundle but thankfully it does not have any “Provided Dependencies” or “Compile Dependencies”. The Jupyter notebooks provided for this blog post include pinned versions for all the necessary dependencies but in case you need to update them, this is the process.
Deploy Helm chart
Configure Helm chart
git clone https://github.com/CptOfEvilMinions/BlogProjects.gitcd k8s-jupyter-spark-minio
Spark

By default the Helm chart will use spark version 3.5.0 (spark.image.tag), with an autoscale policy (spark.worker.autoscaling) to create at least one worker (spark.worker.autoscaling.minReplica) with a maximum worker count of 2 (spark.worker.autoscaling.maxReplica), and each worker can only use 1GB of memory max (spark.worker.memoryLimit). These settings are configurable to your use case but the ones provided are a good starting point.
Jupyterhub

By default the Helm chart will use jupyterhub all-spark-notebook (jupyterhub.singleuser.image.name) which includes Python, R, and spark related components for version 3.5.0 of Spark (jupyterhub.singleuser.image.tag). Lastly, we disable the networkPolicy so that notebooks are able to make network connections without restriction.
Minio

By default the Helm chart will deploy Minio in standalone (minio.mode) mode with a replica count (minio.replicas) of 1 to keep our stack simple. Our Minio service will have a persistence volume that claims (minio.persistence.size) 20Gi of space. The Minio Helm chart also supports creating user accounts upon deployment. Our helm chart will create a user called analyst with the ability to read/write (minio.users.analyst.policy) to all S3 buckets, and it will assign the secret value located in the k8s secret (if configured) to the user account. Lastly, our Helm chart will create buckets for logs and these buckets are named using the Databricks medallion architecture.
Jupyter-driver
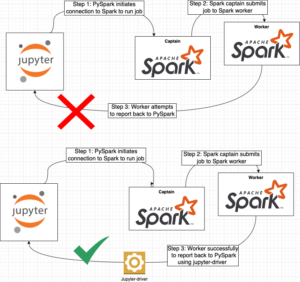
This very small but very important k8s resource deserves its own section. When you start a remote Spark session the worker needs the ability to report its status back to the Jupyter notebook. Unfortunately, reporting the status back to the notebook is performed by the worker initiating a network connection to the Jupyter instance. This is an issue because by default, k8s does not expose the Jupyterhub container in a manner that the worker can connect to it (error below). It took me FOREVER to figure out this is what was happening behind the scenes and the only reason I was able to figure this out is these blog posts Spark – How to fix “WARN TaskSchedulerImpl: Initial job has not accepted any resources” and Spark on Kubernetes: Jupyter and Beyond. The first blog post helped me understand the issue and the second blog helped me understand how to fix the issue. All the credit for the solution to this issue and the clever hack should be given to those authors.
Py4JJavaError: An error occurred while calling None.org.apache.spark.api.java.JavaSparkContext. : java.nio.channels.UnresolvedAddressException at java.base/sun.nio.ch.Net.checkAddress(Net.java:149) at java.base/sun.nio.ch.Net.checkAddress(Net.java:157) at java.base/sun.nio.ch.ServerSocketChannelImpl.netBind(ServerSocketChannelImpl.java:330) at java.base/sun.nio.ch.ServerSocketChannelImpl.bind(ServerSocketChannelImpl.java:294) at io.netty.channel.socket.nio.NioServerSocketChannel.doBind(NioServerSocketChannel.java:141) at io.netty.channel.AbstractChannel$AbstractUnsafe.bind(AbstractChannel.java:562) at io.netty.channel.DefaultChannelPipeline$HeadContext.bind(DefaultChannelPipeline.java:1334) at io.netty.channel.AbstractChannelHandlerContext.invokeBind(AbstractChannelHandlerContext.java:600) at io.netty.channel.AbstractChannelHandlerContext.bind(AbstractChannelHandlerContext.java:579) at io.netty.channel.DefaultChannelPipeline.bind(DefaultChannelPipeline.java:973) at io.netty.channel.AbstractChannel.bind(AbstractChannel.java:260) at io.netty.bootstrap.AbstractBootstrap$2.run(AbstractBootstrap.java:356) at io.netty.util.concurrent.AbstractEventExecutor.runTask(AbstractEventExecutor.java:174) at io.netty.util.concurrent.AbstractEventExecutor.safeExecute(AbstractEventExecutor.java:167) at io.netty.util.concurrent.SingleThreadEventExecutor.runAllTasks(SingleThreadEventExecutor.java:470) at io.netty.channel.nio.NioEventLoop.run(NioEventLoop.java:569) at io.netty.util.concurrent.SingleThreadEventExecutor$4.run(SingleThreadEventExecutor.java:997) at io.netty.util.internal.ThreadExecutorMap$2.run(ThreadExecutorMap.java:74) at io.netty.util.concurrent.FastThreadLocalRunnable.run(FastThreadLocalRunnable.java:30) at java.base/java.lang.Thread.run(Thread.java:833)
Essentially, we create a k8s service that allows access to the Jupyterhub container on a specified port. Additionally, in our Jupyter notebook we specify the name of this k8s service which is really a DNS name to call back and we specify the port the notebook will listen on. All this together allows a PySpark session to be created and successfully utilized. Lastly, it should be noted that this solution is limited and should NOT be used in a multi-tenant environment – the k8s service ONLY allows for a single call back per Jupyter instance
Deploy
- Create a namespace:
kubectl create ns <namespace>- For this blog post I am going to create a namespace called
foxship
- For this blog post I am going to create a namespace called
cp values-example.yaml values.yaml- Create password for minio root user:
kubectl create secret generic minio-root-secret -n <namespace> --from-literal=rootUser=root --from-literal=rootPassword=$(openssl rand -base64 32 | tr -cd '[:alnum:]') - By default the Helm chart will set the
analystuser password and secretKey toanalyst123- Uncomment the following lines and run the following command

- Create password for minio analyst user:
kubectl create secret generic analyst-s3-access-key -n foxship --from-literal=username=analyst --from-literal=password=$(openssl rand -hex 20) --from-literal=secretKey=analyst
- Uncomment the following lines and run the following command
- Deploy Helm chart:
helm install <release name> . -f values.yaml -n foxship
- Watch the stack spin up:
kubectl get pods -n <namespace> -w
Log into JupyterHub
kubectl port-forward -n <namespace> svc/proxy-public 8080:80- Open a web browser to
http://127.0.0.1:8080- Enter
jovyanfor username - Enter
jupyterfor password
- Enter
- Settings > Themes > JupyterHub Dark

- In the top left hand side select the upload icon (upward arrow)
- Find the github repo on your local machine and upload all the notebooks in
k8s-jupyter-spark-minio/notebooks
Jupyterhub Notebook: init.ipynb
Install Python dependencies
!pip3 install --quiet pyspark==3.5.0 !pip3 install --quiet delta-spark==3.0.0 !pip3 install --quiet minio==7.2.0
Download dataset
For this tutorial I am going to download the DEFCON 30 Project Obsidian dataset. This dataset contains logs generated by security tooling in a simulated environment. The important thing to note in this section is that when we initially download the data it exists in our Jupyter lab environment, which is not accessible by Apache Spark workers. We need to upload the log files from disk to Minio (s3), which is done in the next section.
curl -s https://btv.cachefly.net/DC30/Obsidian/obsidian_logs_DC30_v2.02.zip \
--output /tmp/obsidian_logs_DC30_v2.02.zip
unzip -P "obsidian is never going to give you up" /tmp/obsidian_logs_DC30_v2.02.zip
Upload log files to Minio (S3)
This section uses the Minio Python dependency to read logs from the JupyterHub disk to Minio S3 bucket. We start by iterating the environment variables (env) to generate the Minio endpoint so that we can generate the Minio client (client = Minio(). Next, we iterate through the logs on disk (osquery, sysmon, hmail, winlogbeats, and zeek) and upload each log file to a specified S3 bucket and path.
from minio.error import S3Error
from minio import Minio
import os.path
import glob
import os
MINIO_ENDPOINT = str()
for name, value in os.environ.items():
if name.endswith("_MINIO_PORT"):
MINIO_ENDPOINT=value.replace("tcp://", "")
print(MINIO_ENDPOINT)
client = Minio(
MINIO_ENDPOINT,
access_key="analyst-svcacct",
secret_key="analyst123",
secure=False
)
found = client.bucket_exists("fallback")
print(found)
for log_file in glob.glob("filebeat/osquery-*.log"):
client.fput_object(
"logs-bronze",
f"osquery/{os.path.basename(log_file)}",
log_file,
)
print(
log_file, "successfully uploaded as object",
f"logs-bronze/osquery/{os.path.basename(log_file)} to bucket logs-bronze",
)
for log_file in glob.glob("filebeat/sysmon-*.log"):
client.fput_object(
"logs-bronze",
f"sysmon/{os.path.basename(log_file)}",
log_file,
)
print(
log_file, "successfully uploaded as object",
f"logs-bronze/sysmon/{os.path.basename(log_file)} to bucket logs-bronze",
)
for log_file in glob.glob("filebeat/hmail-*.log"):
client.fput_object(
"logs-bronze",
f"hmail/{os.path.basename(log_file)}",
log_file,
)
print(
log_file, "successfully uploaded as object",
f"logs-bronze/hmail/{os.path.basename(log_file)} to bucket logs-bronze",
)
for log_file in glob.glob("filebeat/wineventlogs-*.log"):
client.fput_object(
"logs-bronze",
f"windows/{os.path.basename(log_file)}",
log_file,
)
print(
log_file, "successfully uploaded as object",
f"logs-bronze/wineventlogs/{os.path.basename(log_file)} to bucket logs-bronze",
)
for log_file in glob.glob("zeek/*/*.log"):
client.fput_object(
"logs-bronze",
f"zeek/{os.path.basename(log_file)}",
log_file,
)
print(
log_file, "successfully uploaded as object",
f"logs-bronze/zeek/{os.path.basename(log_file)} to bucket logs-bronze",
)
kubectl port-forward -n <namespace> svc/stack-minio-console 9001:9001- Open a web browser to
http://127.0.0.1:9001- Enter
analystas username - Enter the analyst password
- If you generated a random password run:
kubectl get secret -n <namespace> analyst-s3-access-key -o jsonpath="{.data.password}" | base64 --decode ; echo
- If you generated a random password run:
- Enter
- Object browser > logs-bronze

- Object browser > logs-bronze > osquery

Download dependencies/JARs for Spark
The biggest callout for this section is the amazing capability that PySpark provides to supply a JAR as part of the session. Several tutorials on the internet have you download these JARs ahead of time and bake them into the container images. Trying to create/manage custom images in Docker is easy but with K8s it’s not as straightforward. Thankfully, PySpark has the capability to load JARs to be used at run time. At a high level, we are downloading JARs that will allow us to interact with an S3-like (Minio) system and the ability to use Delta tables.
# find /usr/local/spark/jars/ -name 'hadoop-client-api-*.jar' | awk -F- '{print $4}' | grep -Eo '([0-9]\.)+[0-9]'
export HADOOP_VERSION="3.3.4"
export AWS_VERSION="1.12.599"
export DELTA_VESION="3.0.0"
export SCALA_VERSION="2.12"
curl -s -O https://repo1.maven.org/maven2/org/apache/hadoop/hadoop-common/${HADOOP_VERSION}/hadoop-common-${HADOOP_VERSION}.jar
curl -s -O https://repo1.maven.org/maven2/org/apache/hadoop/hadoop-aws/${HADOOP_VERSION}/hadoop-aws-${HADOOP_VERSION}.jar
curl -s -O https://repo1.maven.org/maven2/com/amazonaws/aws-java-sdk-core/${AWS_VERSION}/aws-java-sdk-core-${AWS_VERSION}.jar
curl -s -O https://repo1.maven.org/maven2/com/amazonaws/aws-java-sdk-s3/${AWS_VERSION}/aws-java-sdk-s3-${AWS_VERSION}.jar
curl -s -O https://repo1.maven.org/maven2/com/amazonaws/aws-java-sdk-dynamodb/${AWS_VERSION}/aws-java-sdk-dynamodb-${AWS_VERSION}.jar
curl -s -O https://repo1.maven.org/maven2/io/delta/delta-spark_${SCALA_VERSION}/${DELTA_VESION}/delta-spark_${SCALA_VERSION}-${DELTA_VESION}.jar
curl -s -O https://repo1.maven.org/maven2/io/delta/delta-storage/${DELTA_VESION}/delta-storage-${DELTA_VESION}.jar
Jupyterhub Notebook: osquery-delta.ipynb
Create PySpark session
Like all Python scripts, we start by importing the necessary Python libraries to connect and interact with Spark using Delta tables. The notebook also defines a list of JARs (external_jars) by file path in a list that will load JARs from disk into our PySpark session. Next, we iterate the environment variables (env) to generate the Spark and Minio endpoints so that we can instruct PySpark how to connect to these services. Next, we create a Spark config using the PySpark builder where we define Spark and S3 settings. The Spark settings allow PySpark to interact with the remote Spark cluster. The S3 settings allow PySpark to create/modify files from an S3 compatible object store, in our case Minio. Lastly, we use that PySpark config to create a PySpark session with Delta table functionality.
from delta import *
import pyspark
import os
external_jars = [
"hadoop-aws-3.3.4.jar",
"aws-java-sdk-core-1.12.599.jar",
"aws-java-sdk-s3-1.12.599.jar",
"delta-storage-3.0.0.jar",
"delta-spark_2.12-3.0.0.jar",
"aws-java-sdk-dynamodb-1.12.599.jar",
"hadoop-common-3.3.4.jar",
]
SPARK_ENDPOINT = str()
for name, value in os.environ.items():
if name.endswith("SPARK_MASTER_SVC_SERVICE_HOST"):
SPARK_ENDPOINT=f"spark://{value}:7077"
print(f"SPARK_ENDPOINT: {SPARK_ENDPOINT}")
MINIO_ENDPOINT = str()
for name, value in os.environ.items():
if name.endswith("_MINIO_PORT"):
MINIO_ENDPOINT=value.replace("tcp://", "")
print(f"MINIO_ENDPOINT: {MINIO_ENDPOINT}")
builder = (
pyspark.sql.SparkSession.builder.appName("Myapp")
# Sets the Spark master/captain URL to connect too.
.master(SPARK_ENDPOINT)
# JARs on disk to load into our Spark session
.config("spark.jars", ",".join(external_jars))
# k8s service for Jupyter driver
.config("spark.driver.host", "jupyter-driver")
# Port for Jupyter driver
.config("spark.driver.port", 2222)
# Extending the capabilities of SQL searching with Delta tables
.config("spark.sql.extensions", "io.delta.sql.DeltaSparkSessionExtension")
.config("spark.sql.catalog.spark_catalog", "org.apache.spark.sql.delta.catalog.DeltaCatalog")
####### AWS setup and creds #######
.config("spark.hadoop.fs.s3a.aws.credentials.provider", "org.apache.hadoop.fs.s3a.SimpleAWSCredentialsProvider")
.config("spark.hadoop.fs.s3a.access.key", "analyst")
.config("spark.hadoop.fs.s3a.secret.key", "analyst123")
.config("spark.hadoop.fs.s3a.endpoint", MINIO_ENDPOINT)
.config("spark.hadoop.fs.s3a.connection.ssl.enabled", "false")
.config("spark.hadoop.fs.s3a.path.style.access", "true")
.config("spark.hadoop.fs.s3a.attempts.maximum", "1")
.config("spark.hadoop.fs.s3a.connection.establish.timeout", "5000")
.config("spark.hadoop.fs.s3a.connection.timeout", "10000")
)
spark = configure_spark_with_delta_pip(builder).getOrCreate()
print ("done")
Spark session builder breakdown:
.appName()– This option is on the nose with how it works. This will supply a custom name to Spark for naming jobs.master()– Sets the Spark master URL to connect to. In our case, we are specifying a remote Spark master/captain node running on k8s..config("spark.jars": [])– This parameter can take a myriad of options to configure the session. In our case, we provide a list of file paths to JARs on disk to load into our Spark session..config("spark.driver.host", "jupyter-driver")– Specifies the name of the k8s service that we created to allow access to the Jupyterhub container from Spark worker.config("spark.driver.port", 2222)– Specifies the port of the k8s service that we created to allow access to the Jupyterhub container from Spark worker.config("spark.sql.extensions", "io.delta.sql.DeltaSparkSessionExtension")–SparkSessionExtensionsis an Injection API for Spark SQL developers to extend the capabilities of a SparkSession. In our case, we are extending the capabilities of SQL searching with Delta tables..config("spark.sql.catalog.spark_catalog", "org.apache.spark.sql.delta.catalog.DeltaCatalog")–SessionCatalogis a catalog of relational entities in SparkSession (e.g. databases, tables, views, partitions, and functions). In our case, we are extending the capabilities by using Delta as a catalog..config("spark.hadoop.fs.s3a.impl", "org.apache.hadoop.fs.s3a.S3AFileSystem")– The implementation class of the S3A Filesystem.config("spark.hadoop.fs.s3a.aws.credentials.provider", "")– Specify how credentials to access the S3 bucket will be provided. In our case we will be providing access key and secret key.config("spark.hadoop.fs.s3a.access.key", "")– Specify the access key for S3.config("spark.hadoop.fs.s3a.secret.key", "")– Specify the secret key for S3
.config("spark.hadoop.fs.s3a.endpoint", "")– Specify the AWS S3 endpoint to connect to. In our case we will be point this value at Minio.config("spark.hadoop.fs.s3a.connection.ssl.enabled", "")– Specify whether to use SSL/TLS when connecting to the S3 endpoint.config("spark.hadoop.fs.s3a.path.style.access","")– Enable S3 path style access. When using AWS s3 the bucket name is part of the domain and the URI specifies the file path to the location of the object in S3. By setting this value totruewe specify the bucket name in the S3 URL.- AWS S3 object URL:
http://test_bucket.s3.amazonaws.com/abc123_object - Minio object URL:
http://minio.example.local/test_bucket/abc123_object
- AWS S3 object URL:
.config("spark.hadoop.fs.s3a.attempts.maximum", "")– How many times we should retry commands on transient errors.config("spark.hadoop.fs.s3a.connection.establish.timeout", "")– Socket connection setup timeout in milliseconds.config("spark.hadoop.fs.s3a.connection.timeout", "")– Socket connection timeout in milliseconds
Osquery bronze schema
This section of the Jupyterhub notebook ingests the raw Osquery logs from Minio/S3 into a PySpark schema. At a high level each key-value pair of the JSON log has a schema mapping. Breaking down each part of the schema is outside the scope of this blog post. However, to learn more about generating PySpark schemas I recommend these resources: How to Define Schema in Spark Dataframe | Using StructType | Basics of Apache Spark | LearntoSpark and Understanding PySpark’s StructType and StructField for Complex Data Structures. For generating PySpark schemas I recommend using ChatGPT. You can provide it with a JSON blob and ask it to generate a PySpark schema and the accuracy is really good. Keep in mind, that you should not provide any sensitive information to ChatGPT.
from pyspark.sql.types import (
StructField,
StringType,
MapType,
LongType,
IntegerType,
ArrayType,
StructType
)
osquery_bronze_schema = StructType([
StructField("@timestamp",StringType(),True),
StructField("@version",StringType(),True),
StructField('agent', StructType([
StructField('ephemeral_id', StringType(), True),
StructField('hostname', StringType(), True),
StructField('name', StringType(), True),
StructField('type', StringType(), True),
StructField('version', StringType(), True),
])),
StructField('ecs', StructType([
StructField('version', StringType(), True),
])),
StructField('event', StructType([
StructField('dataset', StringType(), True),
StructField('module', StringType(), True),
])),
StructField('fileset', StructType([
StructField('name', StringType(), True),
])),
StructField('host', StructType([
StructField('name', StringType(), True),
])),
StructField('input', StructType([
StructField('type', StringType(), True),
])),
StructField('json', StructType([
StructField('action', StringType(), True),
StructField('calendarTime', StringType(), True),
StructField('columns', MapType(StringType(),StringType()), True),
StructField('counter', IntegerType(), True),
StructField('epoch', LongType(), True),
StructField('unixTime', LongType(), True),
StructField('numerics', IntegerType(), True),
StructField('name', StringType(), True),
StructField('hostIdentifier', StringType(), True),
])),
StructField('log', StructType([
StructField('file', StructType([
StructField('path', StringType(), True),
])),
StructField('offset', LongType(), True),
])),
StructField('service', StructType([
StructField('type', StringType(), True),
])),
StructField("tags",ArrayType(StringType()),True),
])
df = spark.read.schema(osquery_bronze_schema).json("s3a://logs-bronze/osquery/osquery-*log")
df.show()

Drop metadata columns
When Filebeat ingests logs from disk it adds metadata about Filebeat and the log source in addition to the original log. This metadata is not needed for detection and response therefore we are going to instruct Pandas to drop those fields. The Databricks medallion method starts by ingesting the raw log events which is referred to as the bronze phase. Next, we move to the silver phase where we massage the data such as removing unnecessary fields or renaming them.
osquery_bronze = df
drop_metadata_columns = [
"@timestamp",
"@version",
"agent",
"ecs",
"input",
"log",
"tags",
"fileset",
"service",
]
osquery_silver_no_metadata = osquery_bronze.drop(*drop_metadata_columns)
Rename columns and pull data to top level
In this section we are pulling nested JSON fields to the root of the log to make them more accessible.
from pyspark.sql.functions import col
osquery_silver = osquery_silver_no_metadata.select(
col("json.hostIdentifier").alias("hostname"),
col("json.action").alias("action"),
col("json.name").alias("table"),
col("json.unixTime").alias("unixTime"),
col("json.columns").alias("columns"),
)
osquery_silver.show()

Write dataframe as Delta Lake table
Lastly, we need to write our newly modified data to Minio/S3 as a Delta table.
osquery_silver.write.format("delta")\
.mode('overwrite')
.save("s3a://logs-silver/osquery/osquery.delta")

Jupyterhub Notebook: osquery-query.ipynb
Read the delta table into a dataframe
To query our Delta lake table we need to read from Minio/S3 into a Pandas data frame.
df = spark.read.format("delta")\
.load("s3a://logs-silver/osquery/osquery.delta")
Query delta table
Once the Delta lake table is read into a Pandas data frame we can perform Pandas functions on the data. In the example below, we are searching process_events for when cmdline contains the letter b and to return the hostname, cmdline columns from events.
from pyspark.sql import functions as F
df.printSchema()
df.where(df.table == "process_events") \
.select(df.hostname, F.col("columns").getItem("cmdline").alias("cmdline")) \
.where(F.col("columns").getItem("cmdline").contains("b")) \
.show(10, False)

Bonus: Connect VScode to remote Jupyter
- Connect to a remote Jupyter server
- Connect to a JupyterHub from Visual Studio Code
- Working with Python interpreters
- Create local proxy to Jupyterhub:
kubectl port-forward -n <namespace> svc/proxy-public 8080:80 - Open a web browser to
http://127.0.0.1:8080and login - File > Hub Control Panel
- Select “Token” in top left
- Enter a name for token
- Select an expiration

- Select “Request a new API token”

- Open a Jupyter notebook in VScode
- Click “Select kernel” in the top right
- Select “Existing JupyterHub server”
- Enter Jupyter server remote URL

- You may get a prompt to install JupyterHub extension, select “Install”

- You may get a prompt to install JupyterHub extension, select “Install”
- Enter username

- Enter user token

- Name Jupyter server

- Select recommended kernel

- Run Jupyter notebook

Areas for improvement
Migrate dedicated works to k8s workers
With the current setup, we have two dedicated Apache Spark workers that are consuming CPU/memory even when not in use. Apache Spark has adapted to leverage K8s and spin up workers dynamically as k8s pods. This capability means we don’t waste resources when there are no active jobs. However, this capability is not easy to setup and is something I would like to explore.
References
- jupyter/all-spark-notebook
- Download Apache Spark
- Databricks medallion architecture
- DEFCON 30 Project Obsidian dataset
- Project-Obsidian-DC30
- Python | os.path.basename() method
- MinIO Python Client SDK for Amazon S3 Compatible Cloud Storage
- Introduction to PySpark JSON API: Read and Write with Parameters
- DataOps 02: Spawn up Apache Spark infrastructure by using Docker
- helm/jupyterhub/jupyterhub
- helm/bitnami/spark
- Getting Started With Apache Spark, Python and PySpark
- Mount Environment Variables Safely with Kubernetes Secrets and Helm chart
- Spark on Kubernetes: Jupyter and Beyond
- unzip password protected zip in unix
- What is a medallion architecture?
- jupyter/all-spark-notebook
- com/amazonaws/aws-java-sdk-s3/1.11.1026
- com/amazonaws/aws-java-sdk/1.11.1026/aws-java-sdk-1.11.1026.jar
- org/apache/hadoop/hadoop-aws/3.3.2/hadoop-aws-3.3.2.jar
- com/amazonaws/aws-java-sdk-core/1.11.1026/aws-java-sdk-core-1.11.1026.jar
- com/amazonaws/aws-java-sdk-bundle/1.11.1026/aws-java-sdk-bundle-1.11.1026.jar
- com/amazonaws/aws-java-sdk-s3/1.11.1026/aws-java-sdk-s3-1.11.1026.jar
- AWS SDK For Java Bundle » 1.11.1026
- AWS SDK For Java Core » 1.11.1026
- AWS Java SDK :: Services :: Amazon S3 » 2.18.41
- Apache Hadoop Amazon Web Services Support » 3.3.2
- Spark – How to fix “WARN TaskSchedulerImpl: Initial job has not accepted any resources”
- SparkSessionExtensions
- Hadoop-AWS module: Integration with Amazon Web Services
